Data and model code from: Tracing growth patterns in cod (Gadus morhua L.) using bioenergetic modelling
Data files
Oct 16, 2023 version files 11.45 MB
-
energy_dens_and_pi_table.csv
275 B
-
multinom_mod.rds
212.67 KB
-
observer_model.rds
5.47 KB
-
README.md
15.56 KB
-
smalk_raw_data_males.csv
873.57 KB
-
SST_SBT_Tdiff_1979_2018.csv
702.67 KB
-
stomach_model_incl_small.rds
168.47 KB
-
temp_at_3depth_strata_1979_2018.csv
9.45 MB
-
WBC_bioenergetic_growth_model_v1_0.R
26.66 KB
Abstract
Understanding individual growth in commercially exploited fish populations is key to successful stock assessment and informed ecosystem-based fisheries management. Traditionally, growth rates in marine fish are estimated using otolith age-reading in combination with age-length relationships from field samples, or tag-recapture field experiments. However, for some species, otolith-based approaches have been proven unreliable, and tag-recapture experiments suffer from high working effort and costs as well as low recapture rates. An important alternative approach for estimating fish growth is represented by bioenergetic modelling, which, in addition to pure growth estimation, can provide valuable insights into the processes leading to temporal growth changes resulting from environmental and related behavioural changes. We here developed an individual-based bioenergetic model for Western Baltic cod (Gadus morhua), traditionally a commercially important fish species that however collapsed recently and likely suffers from climate change effects. Western Baltic cod is an ideal case study for bioenergetic modelling because of recently gained in-situ process knowledge on spatial distribution and feeding behaviour based on highly resolved data on stomachs and fish distribution. Additionally, physiological processes such as gastric evacuation, consumption, net-conversion efficiency, and metabolic rates have been well studied for cod in laboratory experiments. Our model reliably reproduced seasonal growth patterns observed in the field. Importantly, our bioenergetic modelling approach implementing depth-use patterns and food intake allowed us to explain the potentially detrimental effect summer heat periods have on growth of Western Baltic cod that likely will increasingly occur in the future. Hence our model simulations highlighted a potential mechanism of how warming due to climate change affects the growth of a key species that may apply for similar environments elsewhere.
Here we provide access to the individual-based bioenergetic growth model which is set up to model the growth of cod in ages 2 to 4 (Gadus morhua L.) in the Belt Sea (western part of the Western Baltic Sea) on a daily basis within one year. The model incorporates contemporary in-situ process knowledge on food intake and seasonal- and temperate-related spatial distribution of cod and allows us to identify seasonal growth patterns. The model is written in the statistical and programming environment R.
title: "ReadMe"author: "Steffen Funk"date: '2023-10-16'output: html_document
Background:
Here you get access to an individual-based bioenergetic growth model which is set up to model the growth of cod in ages 2 to 4 (Gadus morhua L.) in the Belt Sea (western part of the Western Baltic Sea) on a daily basis within one year. The model incorporates contemporary in-situ process knowledge on food intake and seasonal- and temperate-related spatial distribution of cod and allows to identify seasonal growth patterns. For a detailed description of the model (i.e., explanation of equations, submodels used etc.) we refer to the corresponding article by Funk et al. (2023).
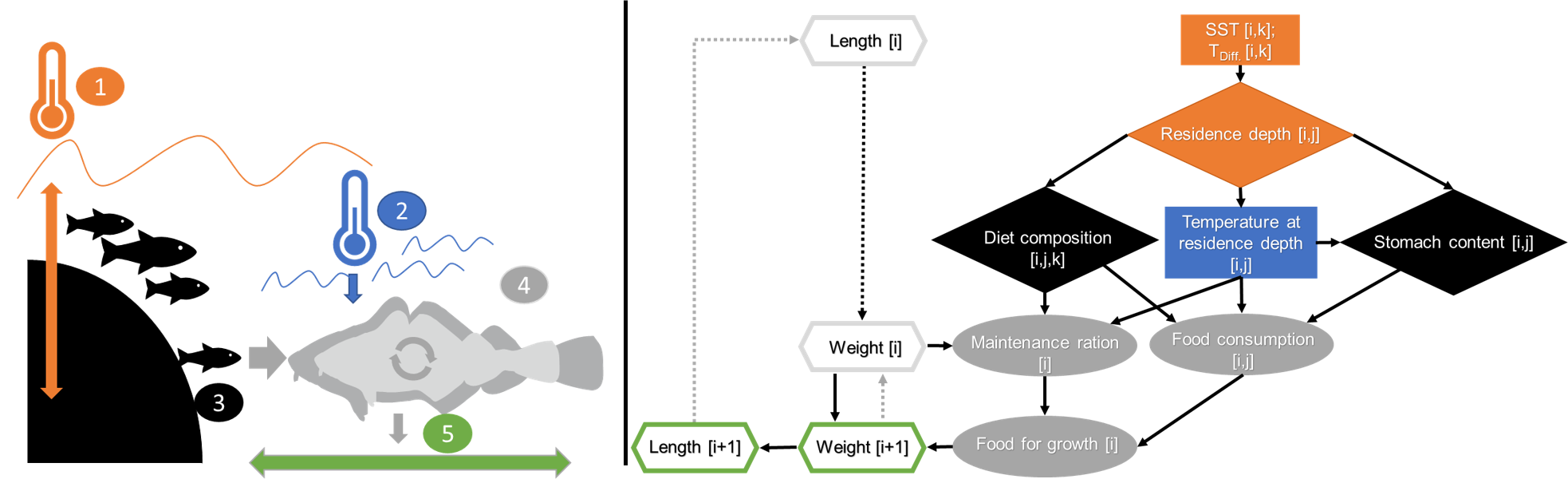 |
|---|
| Set-up of the individual-based bioenergetic growth model. Left panel: Schematic representation of important model components and their influence on growth: (1) temperature conditions (SST = sea surface temperature and TDiff = proxy for stratification, see Material and methods of Funk et al. [currently under review]) (2) temperature at residence depth, (3) food availability and intake, (4) physiological processes, and (5) growth in length. Right panel: Functional pathways (arrows) within the bioenergetic model: rectangles denote temperature variables (SST and TDiff), diamonds denote estimates derived from sub-models I-III; grey ellipses indicate estimates based on published physiological functions; hexagons display cod individual length and weight. i – time step, j – size, k – diet cluster. |
Information on the model script:
The model script contains several functions which are defined in the beginning of the script and corresponding examples which explain how to use the above defined functions.
The bioenergetic growth model relies on several submodels which are imported as rds data files. Furthermore, some data frames (i.e., cod SMALK-data and temperature predictions from the BSIOM) are needed to run the model which can be downloaded as as csv data files and which will be imported when you run the bioenergetic model code.
Note that the bioenergetic growth model for cod in the Western Baltic model was intentionally set up and tested for the modelling periods 2016-2017 and 2016-2017 (since the are the years, where the stomach data was collected). However, we here provide also temperature data for additional years to run the model for other time periods.
The model is written in the statistical and Programming environment R (vers. 4.2.1 "Funny-Looking kid"; R Core Team, 2022)
Before you are getting started:
To run the model script (WBC_bioenergetic_growth_model_v1_0.R), you need to download the following data files:
temp_at_3m_depth_strata_1979_2018.csv (contains predicted temperature values from the BSIOM for 3m depth strata for the period between 1979 and 2018 from the Belt Sea)
SST_SBT_Tdiff_1978_2018.csv (contains predicted sea surface temperature data, sea bottom temperature data and data for the proxy of stratification from BSIOM for the period between 1978 to 2018 from the Belt Sea)
smalk_raw_data_males.csv (contains a dummy data set on biological parameters for cod in the Belt Sea and Sound based on cod biological data recorded during the Baltic international trawl survey which can be downloaded from ICES database DATRAS (ICES BITS)).
energy_dens_and_pi_table.csv (contains a table listing prey specific gastric evacuation coefficients (pi) and energy densities for different prey types (i.e., diet clusters). Values are used for calculation of daily consumption and maintenance ration)
observer_model.rds (contains a fitted linear regression model which is used to calculate the residence depth of a cod. The model is based on a model which explains fishing depth of commercial gill net fishers recorded by at-sea observers by sea surface temperature, a proxy for stratification and the gill net mesh size used. For further details on the model please see also Funk et al., 2020).
stomach_model.rds (contains a fitted generalized additive model which explains stomach content weight of cod with the explanatory variables cod length, temperature at catch depth and catch depth. The model is used to predict the daily stomach contents of cod for a given residence at at a given day. For further details on the model please see also Funk et al., 2021).
multinom.rds (contains a multinomial logistic regression model which explains the diet composition of a cod with the explanatory variables cod length, quarter and catch depth. The model is used to predict the diet composition of cod for a given day. For further details on the model please see also Funk et al., 2021)
Contact information:
For further information, support or help please contact the first author of the study S. Funk (Email: steffen.funk@uni-hamburg.de).
General information:
1. Title of Dataset:
Data and model code from: Tracing growth patterns in cod (Gadus morhua L.) using bioenergetic modelling.
2. Author information:
Funk, Steffen
Universität Hamburg, Hamburg, Germany
contact: steffen.funk@uni-hamburg.de
Funk, Nicole
Universität Hamburg, Hamburg, Germany
Herrmann, Jens-Peter
Universität Hamburg, Hamburg, Germany
Hinrichsen, Hans-Harald
GEOMAR Helmholtz-Institut, Kiel, Germany
Krumme, Uwe
Thünen Institut für Ostseefischerei, Rostock, Germany
Möllmann, Christian
Universität Hamburg, Hamburg, Germany
Temming, Axel
Universität Hamburg, Hamburg, Germany
3. Date of data collection/ code development: 2018-2023
4. Geographic location of the study area: Belt Sea (ICES Subdivision 22, part of the western Baltic Sea)
5. Information about funding sources that supported the development of the growth model:
Bundesministerium für Bildung und Forschung (Germany), Grant no. 03F0914 and Grant no. 03F0863D
Sharing/access information
1. Lincenses/restrictions placed on the data: CC0 1.0 Universal (CC0 1.0) Public Domain
2. Links to publications that cite or use the data:
Funk, S., Funk, N., Herrmann, J.-P., Hinrichsen, H.-H., Krumme, U., Möllmann, C., and Temming. A. (2023).
Tracing growth patterns in cod (Gadus morhua L.) using bioenergetic modelling. Ecology and Evolution.
3. Links to other publicly accessible locations of the data:
https://github.com/FunkSteffen/WBC\_bioenergetic\_growth\_model
4. Links/ relationships to ancillary data sets: None
5. Was data derived from another source? Yes
A. If yes, list source(s): cod single fish data (i.e. SMALK-Data) was retrived from the ICES data repository DATRAS (https://www.ices.dk/data/data-portals/Pages/DATRAS.aspx).
6. Recommended citation for this dataset:
Funk, S., Funk, N., Herrmann, J.-P., Hinrichsen, H.-H., Krumme, U., Möllmann, C., and Temming, A. 2023. Data and model code from: Tracing growth patterns in cod (Gadus morhua L.) using bioenergetic modelling. Dryad Digital Repository. https://doi.org/10.5061/dryad.stqjq2c8r
Data & File overview
1. File list:
A) SST_SBT_Tdiff_1978_2018.csv
B) temp_at_3m_depth_strata_1979_2018.csv
C) smalk_raw_data_males.csv
D) energy_dens_and_pi_table.csv
E) observer_model.rds
F) stomach_model.rds
G) multinom.rds
2. Relationships between files, if important: None
3. Additional related data that was not included in the current data package: None
4. Are there multiple versions of the dataset? No
A. If yes, name of file(s) that was updated: NA
i. Why was the file updated: NA
ii. When was the file updated: NA
DATA-SPECIFIC INFORMATION FOR: temp_at_3m_depth_strata_1979_2018.csv
1. Number of variables: 6
2. Number of cases/rows: 14662
3. Variable List:
julian_day: contains information on the day of the year (Numeric variable from 1 to 365)
month: contains information on the month (Numeric variable from 1 to 12, 1: January, 2: February, 3: March, 4: April, 5: May, 6: June, 7: July, 8: August, 9: September, 10: October, 11: November, 12: December)
year: contans information on the year (Numeric variable from 1979 to 2018)
SBT: contains area-specific mean of modelled sea bottom temperatures retrieved from the Baltic Sea Ice Ocean model for ICES subdivison 22 in °C
SST: contains area specific mean of modelled sea surface temperatures retrieved from the Baltic Sea Ice Ocean model for ICES subdivison 22 in °C
tdiff: contains a proxy for stratificaton in °C computed as the temperature difference between the depth strata 0-3 m (i.e., SST) and the mean of the depth strata 21-27 m.
4. Missing data codes: None
5. Specialized formats or other abbreviations used: None
DATA-SPECIFIC INFORMATION FOR: temp_at_3m_depth_strata_1979_2018.csv
1. Number of variables: 6
2. Number of cases/rows: 233687
3. Variable List:
depth_strat: contains information on the 3 m-depth strata the modelled temperature values belongs to (Factor variable, levels:
1.5 = depth strata 1 to 3 m i.e., surface layer,
4.5 = depth strata 4 to 6m, 7.5 = depth strata 7 to 9,
10.5 = 10-12 m, 13.5 = 13-15 m, 16.5 = 16-18 m, 19.5 = 19-21 m,
22.5 = 22-24 m, 25.5 = 25-27 m, 28.5 = 28-30 m, 31.5 = 31-33 m,
34.5 = 33-36 m, 37.5 = 37-39 m, 40.5 = 40-42 m, 43.5 = 43-45 m,
46.5 = 46-49 m)
julian_day: contains information on the day of the year (Numeric variable from 1 to 365)
month: conteins information on the month of the year (Factor variable, levels:
"jan" = January, "feb" = February, "mar" = March , "apr" = April,
"may" = May, "jun" = June, "jul" = July, "aug" = August, "sep" = September,
"oct" = October, "nov" = November, "dec" = December)
year: contains information on the year. (Numeric variable between 1979 and 2018)
mean_temp: contains a temperature value in °C calculated as an area-specific daily mean for the repective 3 m-depth for the whole SD 22.
depth_strat_new: dummy variable for easier calculation of tdiff (Factor variable which allocates SST to the depth-strata 1-3 m, and SBT to the depth strata 21-23 m and and 24-27 m. For all remaining depth strata the column depth_strat_new contains NA)
4. Missing data codes: NA
5. Specialized formats or other abbreviations used: None
DATA-SPECIFIC INFORMATION FOR: smalk_raw_data_males.csv
1. Number of variables: 16
2. Number of cases/rows: 9725
3. Variable List:
survey: contains information on the survey programme the repective single fish data of cod was collected at (factor variable with only 1 levels: BITS (= Baltic international Trawl survey, monitoring program of the ICES)
year: contains information on the year the data was collected in (Numeric variable between 1992 and 2019)
country: contains information on the country which collected the data (Factor variable with 3 levels: "DEN" = Denmark, "GFR" = Germany, "SWE" = Sweden)
quarter: contains information on the quarter in which the data was collected (Numeric variable. 1 = first quarter)
area: contains information on the ICES subdivision the data was collected in (Factor variable with 2 leves: "22" = ICES subdivison 22 (i.e., Belt Sea), "23" = ICES subdivison 23 (i.e., Sound))
aphia id: list aphia id information of the species for which data was collected (contains only id 126436 which is the id of Alantic cod Gadus morhua L.)
species: scientific name of the species for which data was collected (contains only "Gadus morhua")
length: contains recorded length information in mm of caught cod
plusgr: contains information if the aged cod would be allocated to the plus group for stock assessment prupose i.e., plusgr (contains only missing values i.e. -9)
age: contains information on the cod age obtained from otolith age readings (numeric variable between 0 and 9)
sex: contains information on the sex of the cod (Factor variable, levels: "M" = Male)
maturity: contains information on the maturity stage of cod following the staging criteria of the Baltic international trawl survey (Factor variable, levels:
"1", "3", "2", "5", "4", "-9", "61", "62", "64", "63", "66", "65", "I", "II", "III",
"IV", "V", "VII", "VI", "VIII", "IX"; for further details see ICES, DATRAS)
weight: contains information on the wet weight of cod in g.
canoatlngt: contains information on the number of cod the listed information belongs to (numeric variable between 1 and 11).
dateofcalc: contains information on the date of calculation of the data.
spawning_scale: contains information on the maturity stage of the cod grouped to 7 stages (Factor variable with levels:
"immature" = immature individuals, "spawning" = individuals in spawning stages,
"maturing" = individuals in ripening stages, "resting_skip_spawning" = individuals in resting stages and skip-spawners,
"spent" = spent individuals, "unidentified" = individuals for which no maturty staging was done)
4. Missing data codes: NA, -9, "-9"
5. Specialized formats or other abbreviations used: None
DATA-SPECIFIC INFORMATION FOR: energy_dens_and_pi_table.csv
1. Number of variables: 4
2. Number of cases/rows: 8
3. Variable List:
cluster_name: contains information on the diet cluster the energy density and prey-specific gastrc evacuation coeffiecient belongs to (Factor variable with 8 levels:
Annelids, Clupeids, Common shore crab, Flatfish Molluscs, Other fish, Other crustaceans, Peracarids;
Clusters correspond to diet clusters of cod identified in Funk et al., 2021)
cluster_number: contains infromation on the diet cluster the energy density and prey-specific gastrc evacuation coeffiecient belongs to (Numeric variable between 1 and 8;
1 = Annelids, 2 = Clupeids, 3 = Common shore crab, 4 = Flatfish, 5 = Molluscs,
6 = Other fish, 7 = Other crustaceans, 8 = Peracarids;
Clusters correspond to diet clusters of cod identified in Funk et al., 2021)
pi :contains prey-specific gastric evacuation constants (g * g-(1-α)* g-γ * h^-1) allocated to the respective diet cluster (see Funk et al., 2023 for further details on allocation)
energy_dens:contains energey density in J * g^-1 allocated to the respective diet cluster (see Funk et al., 2023 for further details on allocation)
4. Missing data codes: None
5. Specialized formats or other abbreviations used: None
References:
Funk, S., Funk, N., Herrmann, J.-P., Hinrichsen, H.-H., Krumme, U., Möllmann, C., and Temming. A. **2023.**Tracing growth patterns in cod (Gadus morhua L.) using bioenergetic modelling. Ecology and Evolution.
Funk, S., Frelat, R., Möllmann, C., Temming, A., and Krumme, U. 2021. The forgotten feeding ground: patterns in seasonal and depth-specific food intake of adult cod Gadus morhua in the western Baltic Sea. Journal of Fish Biology, 98(3): 707-722. https://doi.org/10.1111/jfb.14615
Funk, S., Krumme, U., Temming, A., and Möllmann, C. 2020. Gillnet fishers‘ knowledge reveals seasonality in depth and habitat use of cod (Gadus morhua) in the Western Baltic Sea. ICES Journal of Marine Science, 77(5):1816-1829.
ICES, ICES BITS (Baltic International Trawl Survey) dataset. ICES, Copenhagen, Denmark.
R Core Team. 2022. A Language and Environment for Statistical Computing. R foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- Funk, Steffen; Funk, Nicole; Herrmann, Jens‐Peter et al. (2023). Tracing growth patterns in cod (Gadus morhua L.) using bioenergetic modelling. Ecology and Evolution. https://doi.org/10.1002/ece3.10751
